KnoBo incorporates knowledge priors from medical documents via inherently interpretable models.
KnoBo incorporates knowledge priors from medical documents via inherently interpretable models.

While deep networks have achieved broad success in analyzing natural images, when applied to medical scans, they often fail in unexcepted situations. We investigate this challenge and focus on model sensitivity to domain shifts, such as data sampled from different hospitals or data confounded by demographic variables such as sex, race, etc, in the context of chest X-rays and skin lesion images. A key finding we show empirically is that existing visual backbones lack an appropriate prior from the architecture for reliable generalization in these settings. Taking inspiration from medical training, we propose giving deep networks a prior grounded in explicit medical knowledge communicated in natural language. To this end, we introduce Knowledge Bottlenecks (KnoBo), a class of concept bottleneck models that incorporates knowledge priors that constrain it to reason with clinically relevant factors found in medical textbooks or PubMed. KnoBo uses retrieval-augmented language models to design an appropriate concept space paired with an automatic training procedure for recognizing the concept. We evaluate different resources of knowledge and recognition architectures on a broad range of domain shifts across 20 datasets. In our comprehensive evaluation with two imaging modalities, KnoBo outperforms fine-tuned models on confounded datasets by 32.4% on average. Finally, evaluations reveal that PubMed is a promising resource for making medical models less sensitive to domain shift, outperforming other resources on diversity of information and final prediction performance.
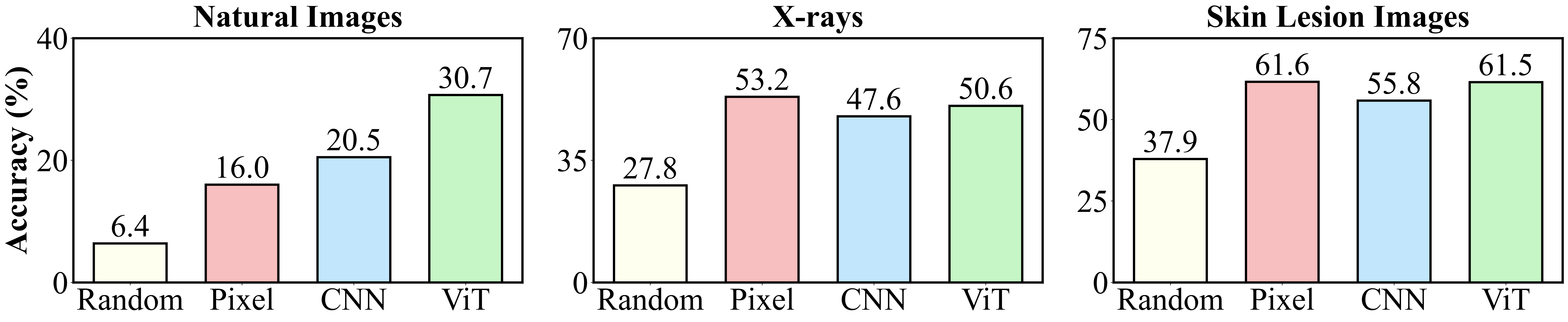
Priors are an important signal allowing models to adopt appropriate hypotheses in low or misleading data regimes. Vision backbones have an effective deep image prior on natural images even when entirely untrained. In contrast, across multiple medical modalities (X-rays and skin lesion images), the deep image prior in current major visual backbones is no more effective than using pixels (and often worse).
Our Knowledge-enhanced Bottlenecks for medical image classification comprises three main components:
(1) Structure Prior constructs the trustworthy knowledge bottleneck by leveraging medical documents;
(2) Bottleneck Predictor grounds the images onto concepts which are used as input for the linear layer;
(3) Parameter Prior constrains the learning of linear layer with parameters predefined by doctors or LLMs.
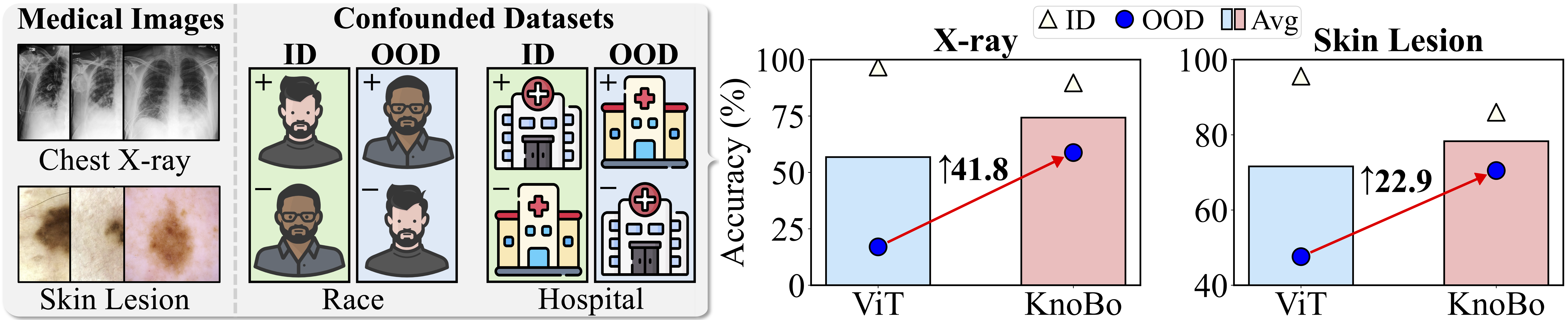
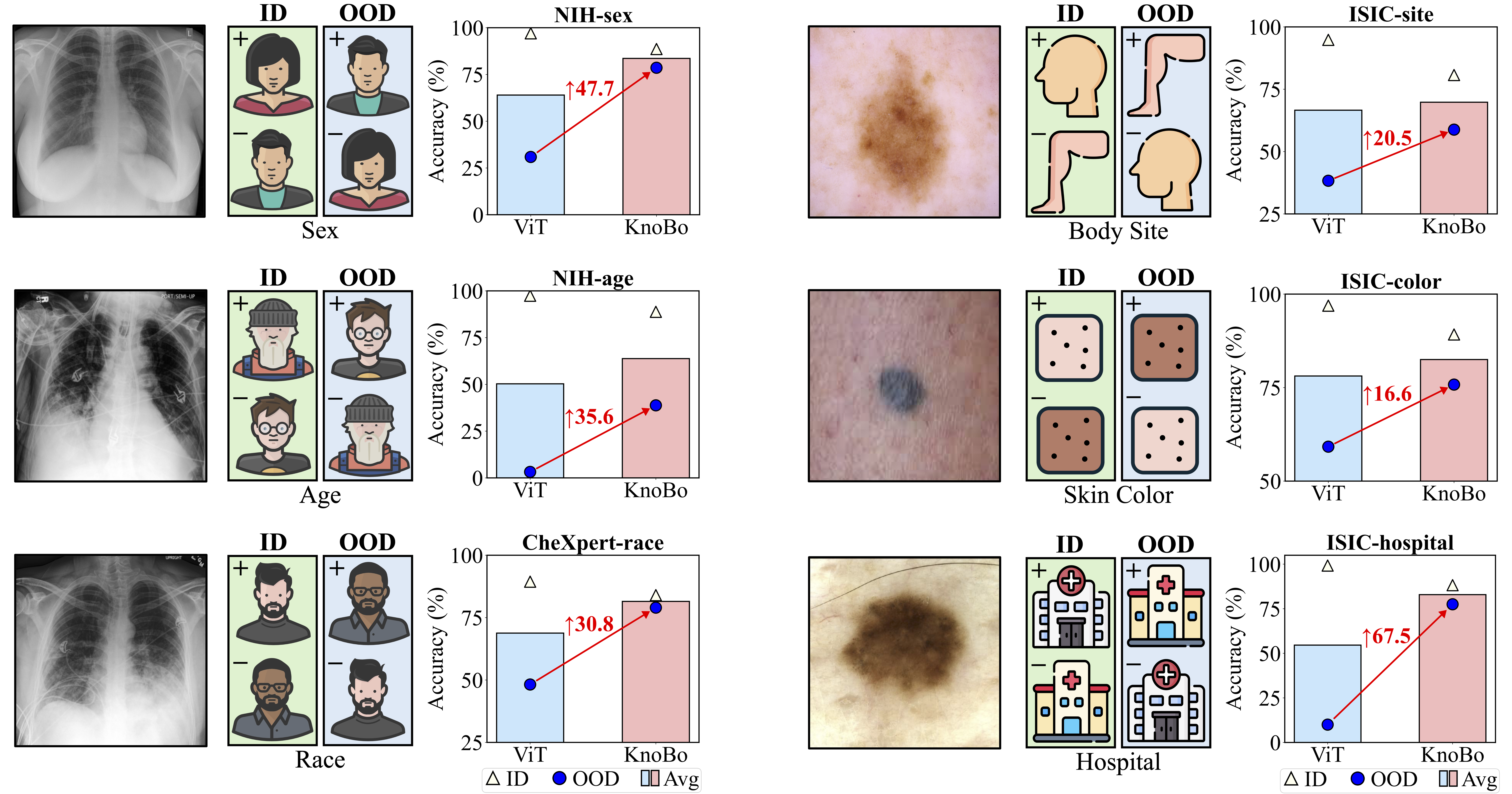
We evaluate 5 confounds per modality, covering scenarios of race, sex, age, scan position, and hospital, etc. Averaged over confounds, KnoBo increases OOD performance by 41.8% and 22.9% on X-ray and skin lesion.
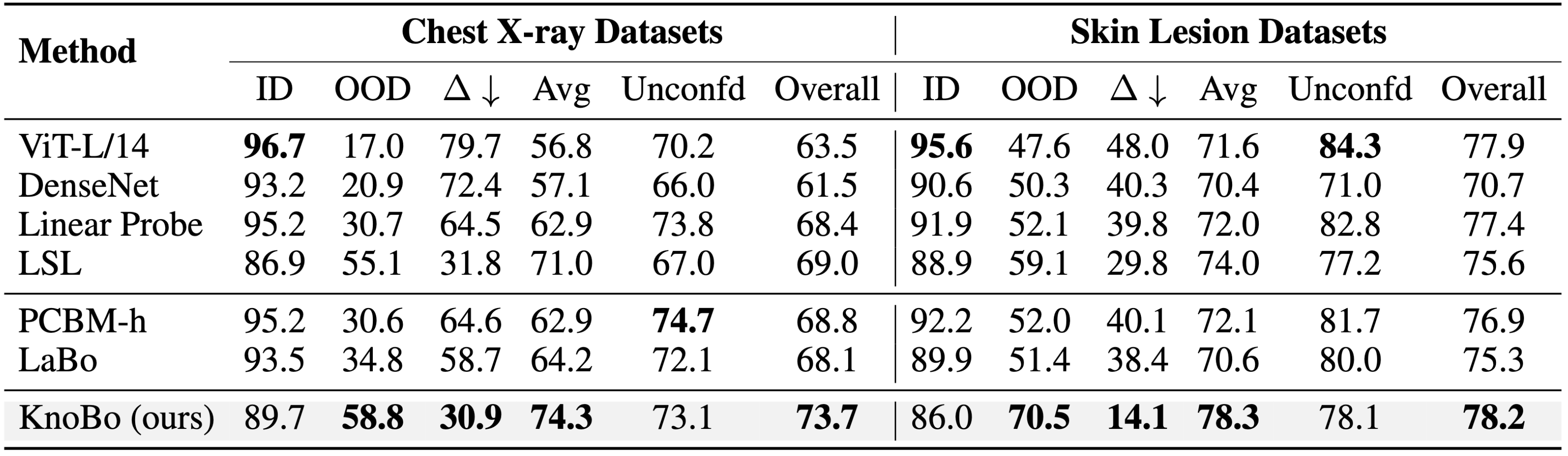
Across 10 confounded and 10 unconfounded medical datasets, KnoBo achieves the best performance on average.
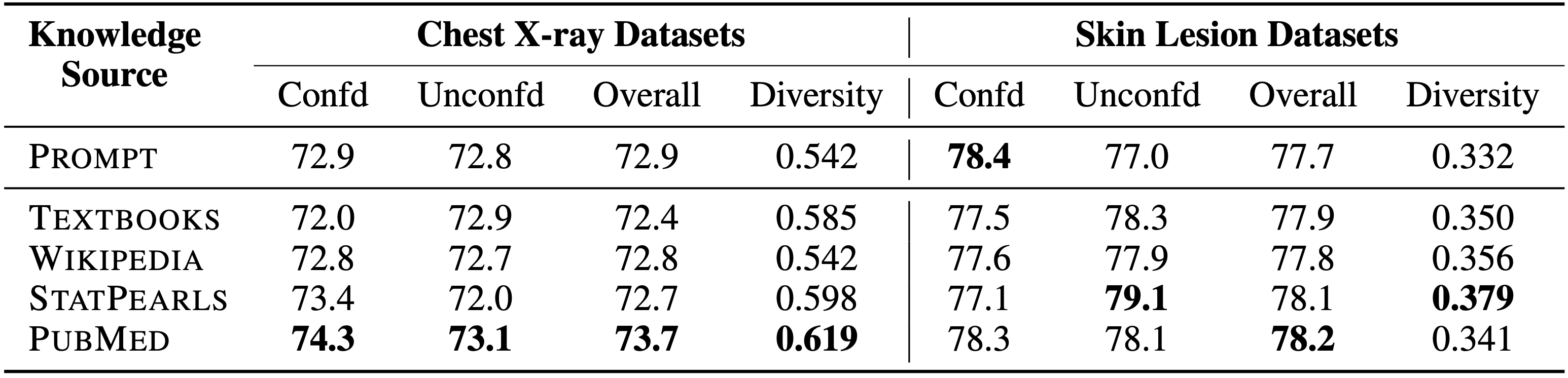
We explore 5 different sources of knowledge for retrieval-augmented generation and reveal that PubMed outperforms other resources in terms of both diversity of information and final prediction performance.
Example concept bottlenecks generated from PubMed:
X-ray Bottleneck (PubMed) Skin Lesion Bottleneck (PubMed)@article{yang2024textbook,
title={A Textbook Remedy for Domain Shifts: Knowledge Priors for Medical Image Analysis},
author={Yue Yang and Mona Gandhi and Yufei Wang and Yifan Wu and Michael S. Yao and Chris Callison-Burch and James C. Gee and Mark Yatskar},
journal={arXiv preprint arXiv:2405.14839},
year={2024}
}